Percent kcal of CARB, PROT, TFAT
Introduction
We will calculate the percentage of calories from each of the three macronutrients in the sum of calories from the three macronutrients. Thus, the percentage of calories from CARB, PROT, and TFAT will add up to 100.
Load functions and packages
Name the path to DietR directory where input files are pulled.
main_wd <- "~/GitHub/DietR"Load necessary packages.
library(ggplot2)Load the necessary functions.
source("lib/specify_data_dir.R")
source("lib/ggplot2themes.R")
source("lib/percent_kcal.R")Load the distinct 100 colors for use.
distinct100colors <- readRDS("lib/distinct100colors.rda")You can come back to the main directory by:
setwd(main_wd)Import data from your data directory
Specify the directory where the data is.
SpecifyDataDirectory(directory.name = "eg_data/NHANES/Laboratory_data/")Load subsetted totals data with males 60-79 years old.
totals <- read.table("QCtotal_d_ga_body_meta_glu_comp_2_males60to79.txt", sep="\t", header=T)Calculate the %kcal of CARB, PROT, and TFAT for each user and take means by Gender_Age
CPTpctKcalPerUser(inputfn=totals, group='GLU_index', across='SEQN',
outfn="QCtotal_d_ga_body_meta_glu_comp_2_M60to79_CPT_kcal_by_GLU.txt")Load the output.
CPT_kcal <- read.table("QCtotal_d_ga_body_meta_glu_comp_2_M60to79_CPT_kcal_by_GLU.txt", sep="\t", header=T)CPT_kcal has Group, macronutrient, n, mean, and sd of each group.
CPT_kcal## Group macronutrient n mean sd
## 1 Diabetic Carbohydrate 62 46.01620 7.478108
## 2 Normal Carbohydrate 47 49.82694 8.938830
## 3 Prediabetic Carbohydrate 127 47.11511 8.516840
## 4 Diabetic Protein 62 16.36385 3.446309
## 5 Normal Protein 47 16.63203 4.070425
## 6 Prediabetic Protein 127 16.37167 3.528409
## 7 Diabetic Total Fat 62 37.61995 7.107032
## 8 Normal Total Fat 47 33.54103 7.651435
## 9 Prediabetic Total Fat 127 36.51322 7.523699Generate a stacked barchart without SD
Order Gender_Age by a certain macronutrient by the “order.by” argument. You can also specify the stacking order of all the macronutrients by the “macronu.order” argument. [Note] The last item will be on the bottom of the barchart.
PlotStackedwoSD(data=CPT_kcal,
order.by = "Carbohydrate",
macronut.order=c("Protein", "Total Fat", "Carbohydrate"))The chart is saved as stacked_wo_SD.
stacked_wo_SD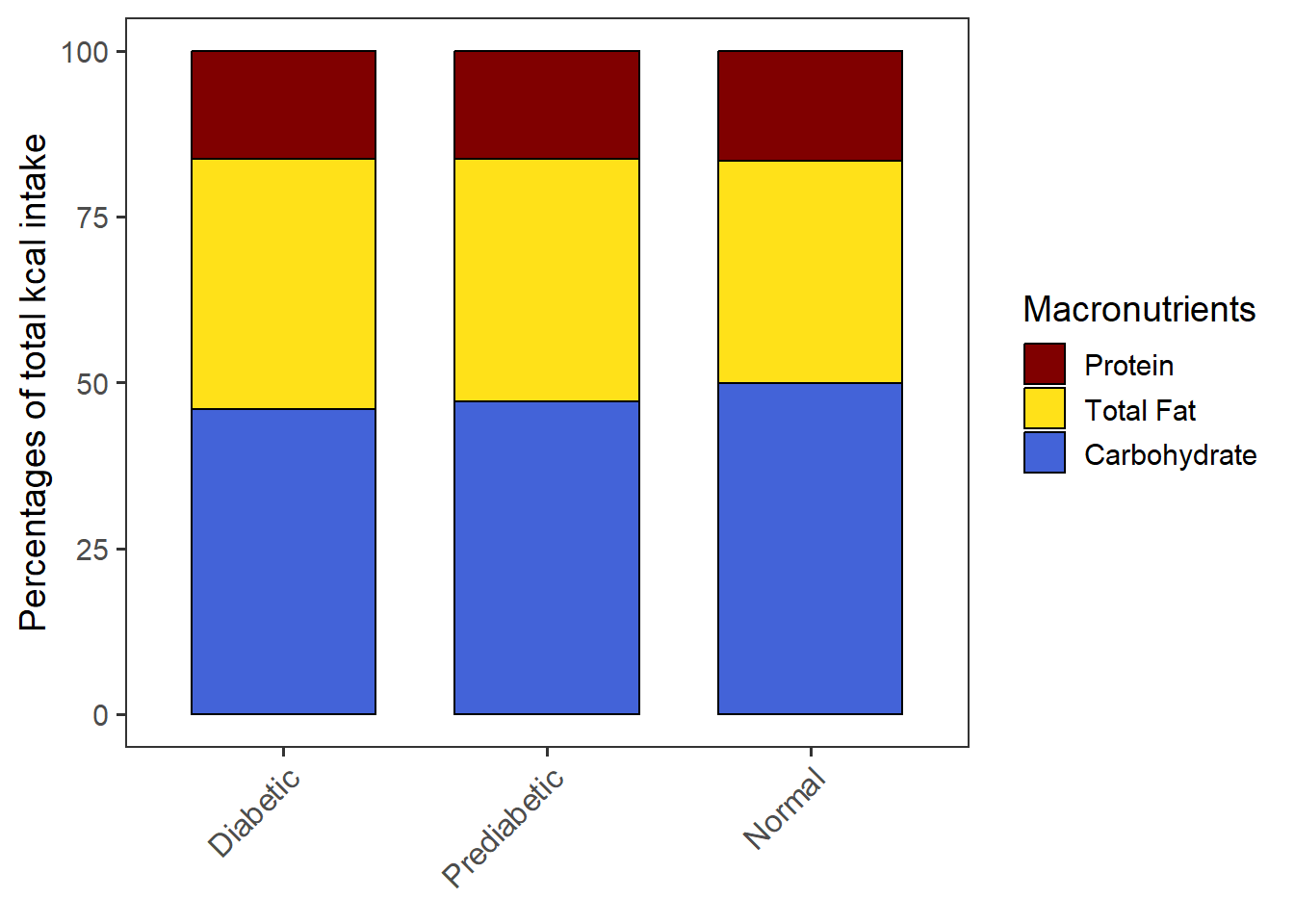
Save as a .pdf.
ggsave("QCtotal_d_ga_body_meta_glu_comp_2_M60to79_CPT_kcal_wo_SD.pdf", stacked_wo_SD,
device="pdf", width=6.2, height=4.3, units="in", dpi=300)When order.by=“NULL”, the diabetic status will be in the alphabetical order by default.
If you want to specify the group order, add the group.order argument.
PlotStackedwoSD(data=CPT_kcal,
order.by = "NULL",
macronut.order=c("Protein", "Total Fat", "Carbohydrate"),
group.order = c("Normal", "Prediabetic", "Diabetic"))stacked_wo_SD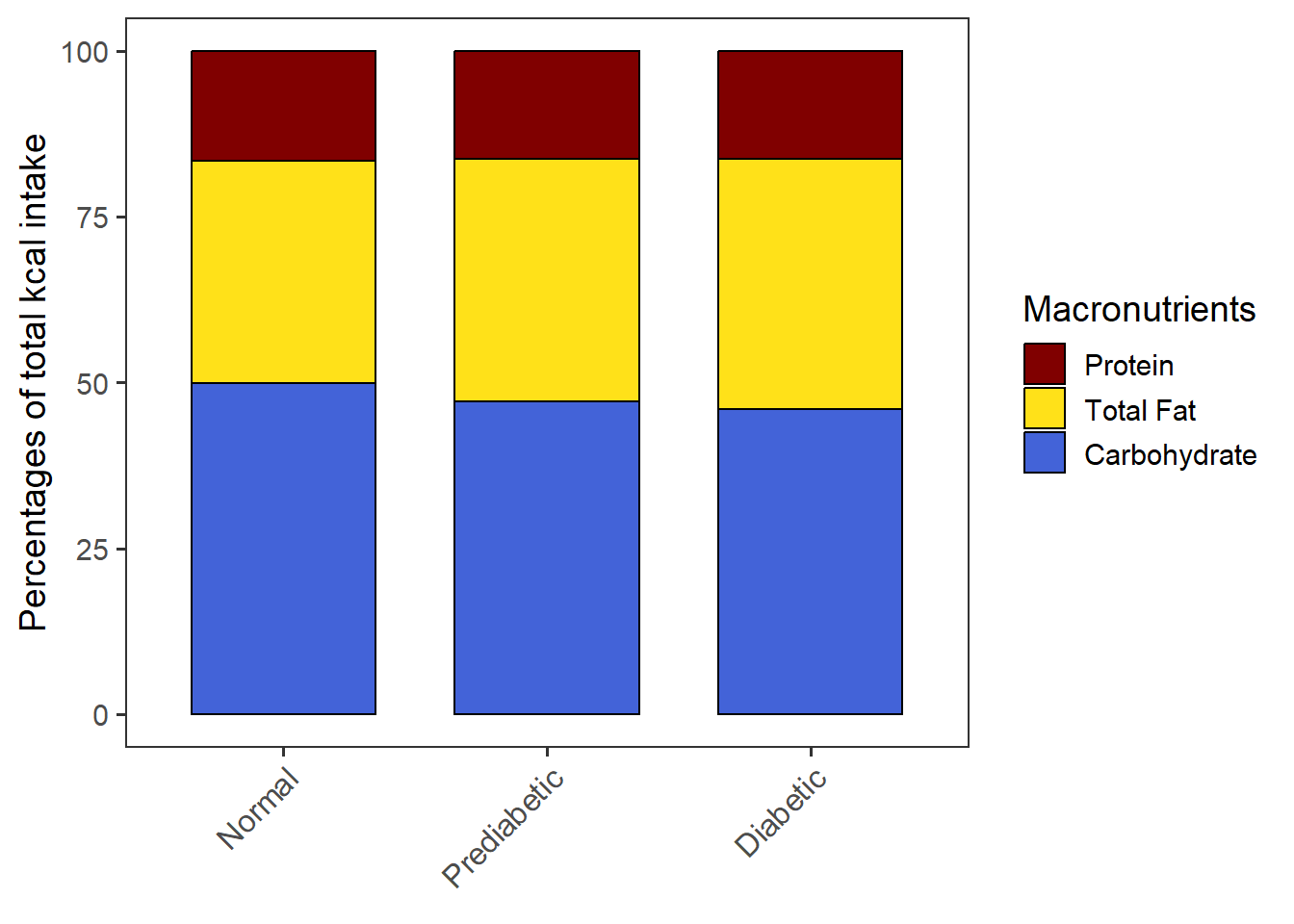
Generate the “dodge”-type of barchart (3 bars per group, NOT STACKED).
Order Diets by a certain macronutrient by the “order.by” argument. You can also specify the plotting order of all the macronutrients by the “macronut.order” argument. Note that the first item will be the leftmost bar.
PlotDodged(data= CPT_kcal,
order.by = "Carbohydrate",
macronut.order=c("Protein", "Total Fat", "Carbohydrate"))The chart is saved as dodged_w_SD.
dodged_w_SD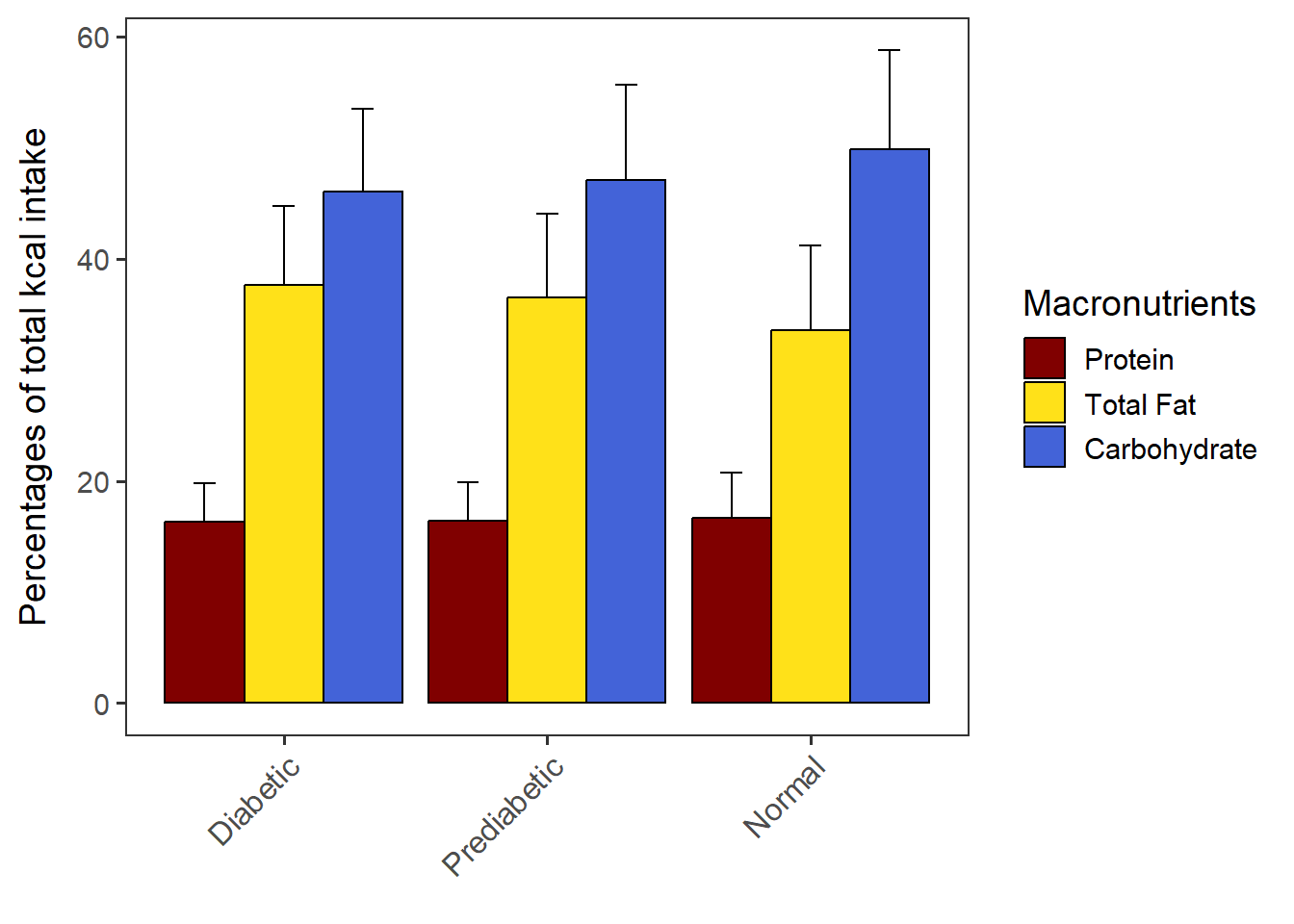
Save it as a .pdf.
ggsave("QCtotal_d_ga_body_meta_glu_comp_2_M60to79_CPT_kcal_dodged_w_SD_NotOrdered.pdf", dodged_w_SD,
device="pdf", width=6.2, height=4, units="in", dpi=300)When order.by=“NULL”, the diabetic status will be in the alphabetical order by default. If you want to specify the group order, add the group.order argument.
PlotDodged(data= CPT_kcal,
order.by = "NULL",
macronut.order=c("Protein", "Total Fat", "Carbohydrate"),
group.order = c("Normal", "Prediabetic", "Diabetic"))dodged_w_SD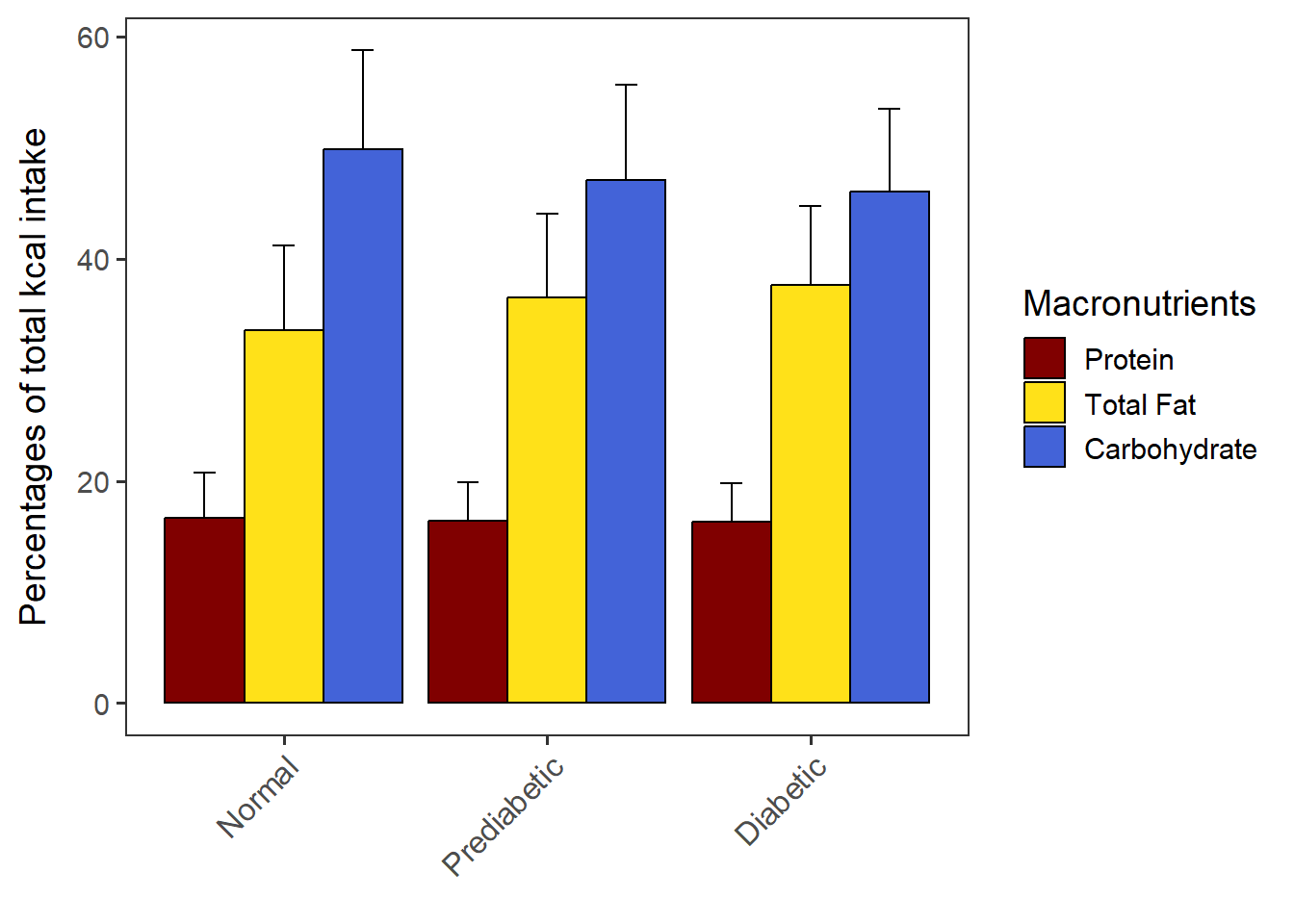
Generate a stacked barchart with SD as error bars
Create a vector that contains all the group levels (Gender_Age, in this case). This “groups” vector will be used in the CalcStackedSD function within the PlotStackedWithSD function. Use the order function to sort the levels in the alphanumeric order.
groups <- unique(CPT_kcal$Group)[order(unique(CPT_kcal$Group))]Order Diet by a certain macronutrient by the “order.by” argument. You can also specify the stacking order of all the macronutrients by the “macronu.order” argument. Note that the last item will be on the bottom of the barchart.
PlotStackedWithSD(data= CPT_kcal,
order.by = "Carbohydrate",
macronut.order= c("Protein", "Total Fat", "Carbohydrate"))The chart is saved as stacked_with_SD.
stacked_with_SD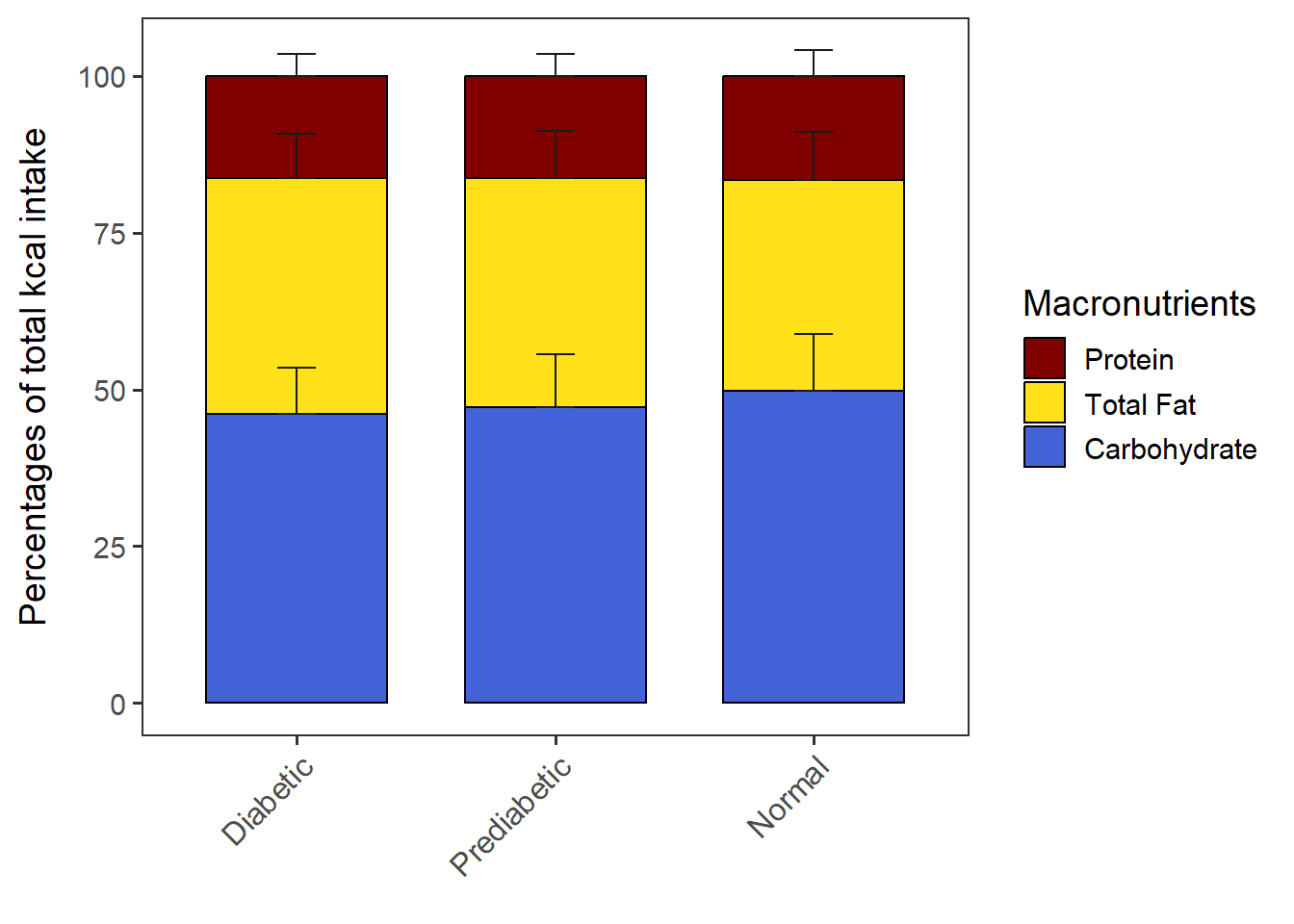
Save as a .pdf.
ggsave("QCtotal_d_ga_body_meta_glu_comp_2_M60to79_CPT_kcal_with_SD.pdf", stacked_with_SD,
device="pdf", width=6.2, height=4.3, units="in", dpi=300)When order.by=“NULL”, the diabetic status will be in the alphabetical order by default. If you want to specify the group order, add the group.order argument.
PlotStackedWithSD(data= CPT_kcal,
order.by = "NULL",
macronut.order= c("Protein", "Total Fat", "Carbohydrate"),
group.order = c("Normal", "Prediabetic", "Diabetic"))stacked_with_SD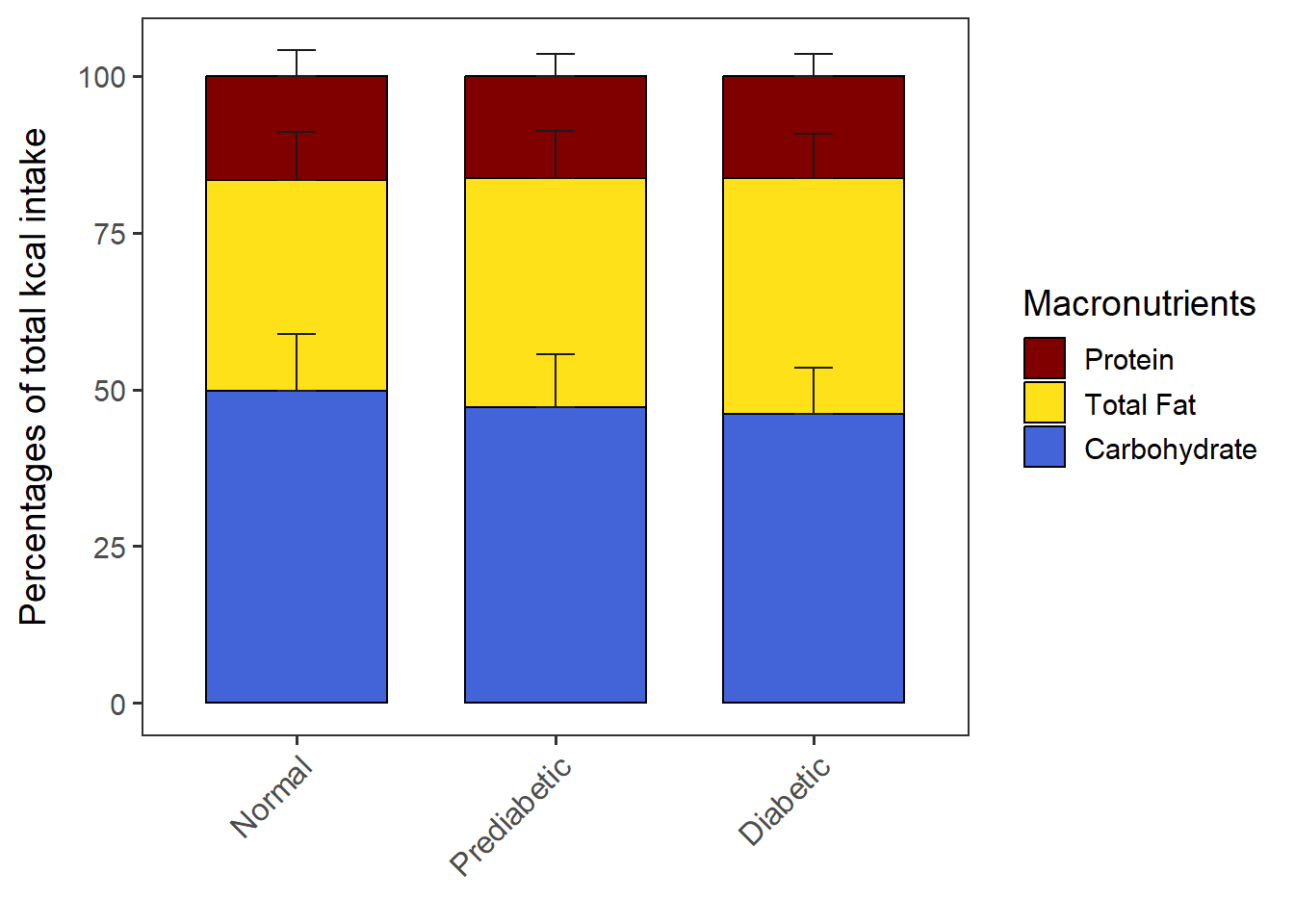
Change the Y axis scale if necessary. Note that if the error bars of Carbohydrates disappear after changing the limits of Y axis, it may be because the error bars are higher than the max Y. Ensure you have enough max Y value to accommodate the error bars.
You can also change the breakpoints of the Y axis.
stacked_with_SD + scale_y_continuous(breaks = c(0, 20, 40, 60, 80, 100))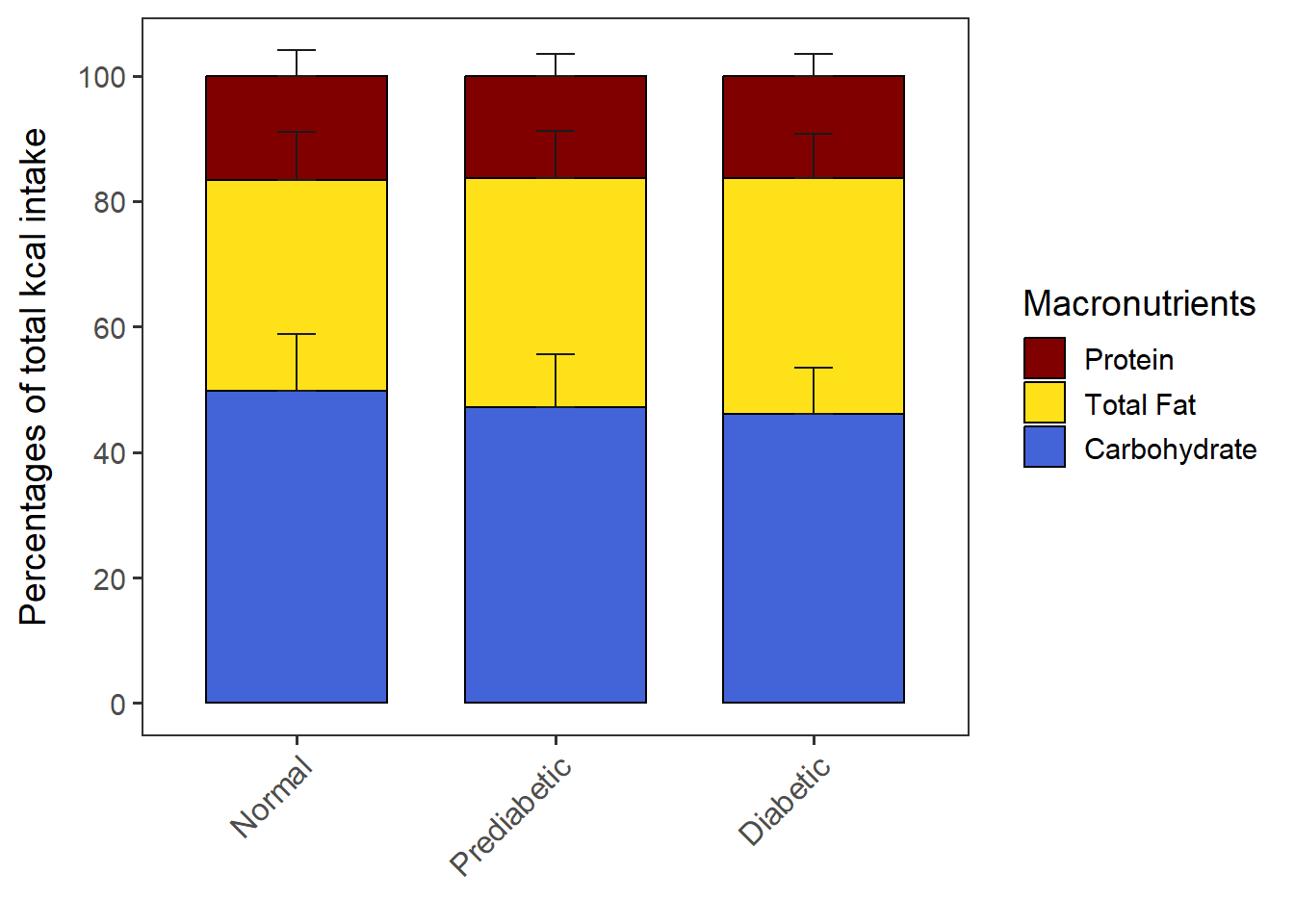
Come back to the main directory.
setwd(main_wd)